Introduction
BiNoM ( Biological Network Manager ) is a Cytoscape plugin developed by the
Computational Systems Biology of Cancer group in
Bioinformatics Laboratory of Institut Curie (Paris).
BiNoM is a Cytoscape plugin,
developed to facilitate the manipulation of biological networks
represented in standard systems biology formats (SBML, SBGN, BioPAX) and to carry out
studies on the network structure. BiNoM provides the user with a
complete interface for the analysis of biological networks in
Cytoscape environment.
BiNoM software supports the following functions:
- Import of BioPAX, SBML and CellDesigner formats
- Export to BioPAX, SBML and CellDesigner formats after user manipulations
- Conversion between standards (CellDesigner->BioPAX, BioPAX->SBML)
- Full support of BioPAX information (reaction network, interaction network, pathway structure, references),
concept of BioPAX index and network interfaces
- Browsing, editing, extracting parts, merging BioPAX files with network graph interface
- Structural analysis of the networks (strongly connected components, path and cycle analysis, network clustering, etc.)
- Support of generating network modular view
- BioPAX network query system: allows to work with huge BioPAX files (such as whole Reactome or NetPath)
- Some general purpose utilities not yet implemented in Cytoscape (clipboard operations, network updating, etc.)
Downloads
BiNoM can be installed and used in several ways.
Using the Cytoscape plugin manager
- Launch the Cytoscape plugin manager (menu "Plugins -> Manage Plugins").
- Go to the category "Other".
- Select the latest version of BiNoM (currently 2.3).
- Click "Install".
- Restart Cytoscape.
As a Cytoscape plug-in file with all libraries included
Installation procedure: just copy the BiNoM jar file into the 'plugin/' folder of Cytoscape (you should also remove all previous BiNoM versions from this folder).
Important note: This version can be used only with versions 2.7.x and 2.8.x of Cytoscape, it not yet compatible with Cytoscape version 3.0.
If you use Cytoscape version 2.4.* or below, you have to follow these steps:
- Download the old version of BiNoM.
- Remove (or, better, rename and keep)
xercesImpl.jar from 'lib/' folder of Cytoscape (since it is not compatible with the version of the same
jar used by Jena in BiNoM).
Important note: Some browsers automatically rename the BiNoM_all.jar to BiNoM_all.zip
after the download. Do not unpack the .zip file but rather rename it back to BiNoM_all.jar and copy into the plugin/ Cytoscape folder.
Using BiNoM as a java library
BiNoM can be linked as a stand-alone java library for file conversions, structural analysis, etc. (examples)
Articles, documentation and goodies
- Bonnet E., Calzone L., Rovera D., Stoll G.,
Barillot E. and Zinovyev A.. BiNoM 2.0, a Cytoscape plugin for accessing
and analyzing pathways using standard systems biology formats. 2013, BMC
Systems Biology, 7:18. web link
- Bonnet, E. Standards and formats in systems biology. Seminar "Mathematics
and Biology of Cancer", 12.06.2013, Institut Curie. slides
- Bonnet, E. BiNoM 2.0, a versatile tool for accessing and analyzing pathways
using standard systems biology formats. NUS - IBENS Workshop "Novel
genome-wide approaches to decipher transcriptional and epigenetic
regulation in mammalian cells. 30 May 2013. ENS, Paris, France. slides
- Cohen, D. BiNoM and NaviCell: Tools for navigation, curation, maintenance
and analysis of complex molecular interaction maps - The 6th International
Biocuration Conference, Cambridge, UK, April 8 2013. slides
- Bonnet, E. BiNoM, a Cytoscape plugin for accessing and analyzing pathways
using standard systems biology formats. Computational Modeling in Biology
Network (COMBINE) 2012. August 2012. The Donnelly Centre, University of
Toronto, Canada slides
- Zinovyev A., Viara E., Calzone L., Barillot E. BiNoM: a Cytoscape plugin
for manipulating and analyzing biological networks. 2008. Bioinformatics
24(6):876-877 manuscript
- BiNoM presentation at the ICSB-2008
workshop Web-services in
Systems Biology (Goteborg, Sweden, August 2008) (slides)
- BiNoM poster presentation at the International Conference on Systems
Biology (Long Beach, CA, October, 2007) (extended abstract)(poster)
Screenshots
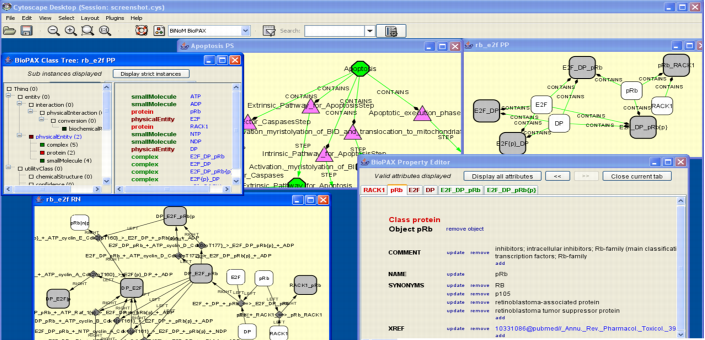 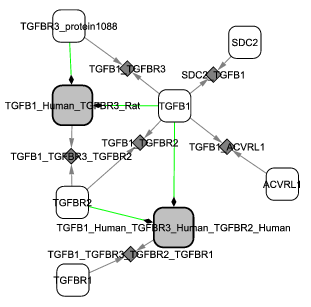
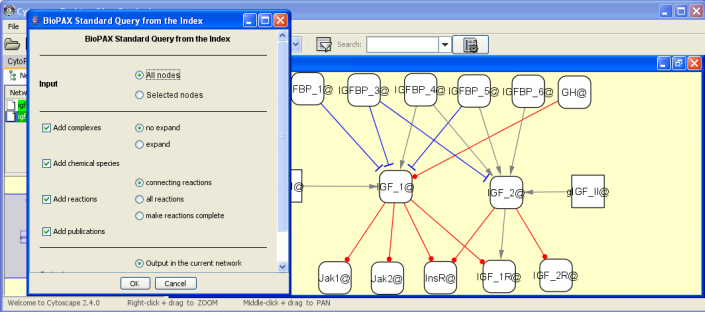
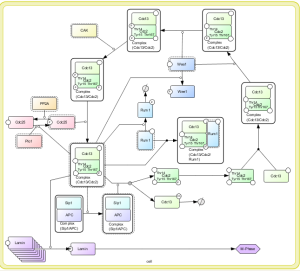 
BiNoM working group
- Andrei Zinovyev
- Eric Bonnet
- Laurence Calzone
- Daniel Rovera
- Gautier Stoll
- Paola Vera-Licona
- Stuart Pook
- Eric Viara
- Emmanuel Barillot
Contacts
Acknowledgements
This project was partly funded by the EC contract ESBIC-D
(LSHG-CT-2005-518192), the PIC Bioinformatique et Biostatistiques
from Institut Curie, the Research Networks Program in
Bioinformatics from the High Council for Scientific and
Technological Cooperation between France and Israel, and the APO-SYS FP7 European program.
|